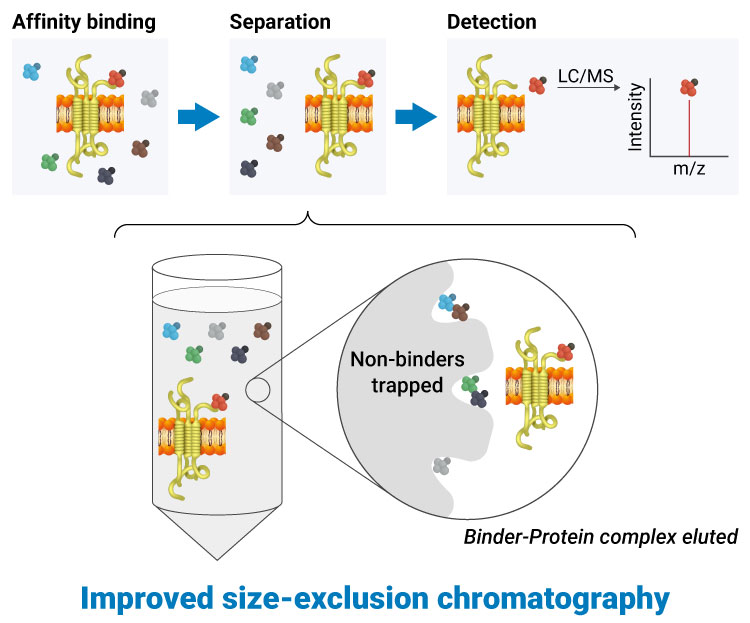
Precise and Versatile Screening with Enhanced ASMS
Our proprietary Binder Selection Technology is a breakthrough evolution of Affinity Selection Mass Spectrometry (ASMS), enabling efficient discovery of specific small-molecule binders even for challenging targets.
In this platform, the process is elegantly simple yet powerful:
Unlike conventional ASMS limited to soluble proteins, our proprietary method extends seamlessly to membrane proteins without requiring solubilization. This allows targets like GPCRs, ion channels, transporters, and even target RNAs to be screened in their native-like conformations. Using a single, streamlined workflow applicable to any target class—including orphan receptors or proteins with unknown function—we efficiently generate highly selective binders. Our extensive screening campaigns have resulted in a growing library of binders against diverse targets, establishing this technology as a versatile and robust solution for next-generation drug discovery.